
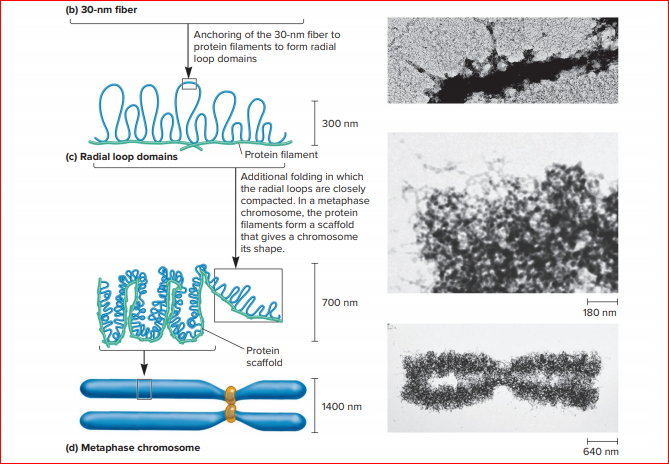

In this model, condensin complexes extrude loops of chromatin through the centre of their ring-like structures ( Cheng et al., 2015, Goloborodko et al., 2016 Ganji et al., 2018 Gibcus et al., 2018). The ‘loop extrusion model’ provides the likely mechanism underlying the formation of mitotic chromosome loops. This radial loop model is supported by early cytological and biochemical observations as well as by more recent Hi-C approaches, which in mitosis detect uniform interaction patterns along chromosomes that are consistent with sequence-independent chromatin loops of between 80 and 120 kb ( Paulson and Laemmli, 1977 Marsden and Laemmli, 1979 Maeshima and Laemmli, 2003, Woodcock and Ghosh, 2010, Naumova et al., 2013 Gibcus et al., 2018). A leading model for mitotic chromosome compaction proposes that chromatin is organised into loops that radiate out from a core scaffold composed of these key organising complexes ( Maeshima and Eltsov, 2008). Key to mitotic chromosome compaction are the action of the condensin complexes, condensin I and condensin II, and topoisomerase IIα, whose depletion or deletion can result in defective chromosome compaction and genome stability in a wide range of organisms ( Hirano, 2012 Maeshima and Laemmli, 2003 Strunnikov et al., 1995, Saka et al., 1994, Wignall et al., 2003 Ono et al., 2003 Hirota et al., 2004 Martin et al., 2016 Woodward et al., 2016 DiNardo et al., 1984 Uemura et al., 1987 Holm et al., 1985). Introductionĭuring mitosis, chromatin is dramatically reorganised and compacted to form individual rod-shaped mitotic chromosomes that allow for proper segregation of the genetic material ( Belmont, 2006). We conclude that heterochromatin alters chromatin loop size, thus contributing to the distinct appearance of heterochromatin on mitotic chromosomes. Hi-C and microscopy indicate that the heterochromatinised fission yeast DNA is organised into smaller chromatin loops than flanking euchromatic mouse chromatin. Here, we show that a similar distinct structure is common to a large subset of insertion events in both mouse and human cells and is coincident with the presence of high levels of heterochromatic H3 lysine nine trimethylation (H3K9me3). A large region of fission yeast DNA inserted into a mouse chromosome was previously observed to adopt a mitotic organisation distinct from that of surrounding mouse DNA. Condensin complexes are involved in chromatin compaction, but the contribution of other chromatin proteins, DNA sequence and histone modifications is less understood. During mitosis chromosomes reorganise into highly compact, rod-shaped forms, thought to consist of consecutive chromatin loops around a central protein scaffold.


 0 kommentar(er)
0 kommentar(er)
